Superstable Elements in Human Genomes
I. What is Superstable elements in human genomes?
Superstable elements are sequences that share 100% identity with no insertions or deletions among multiple human genomes.
II. Human genome data sources
|
Population |
Grouping |
Individuals |
Sequencing Method |
|
European |
Individual |
J. Craig Venter1 |
Sanger Dideoxy |
|
Mother, father, and child trio2 |
NA12878 |
ILLUMINA, LS454, SOLID |
|
|
NA12890 |
ILLUMINA |
||
|
NA12892 |
ILLUMINA |
||
|
West African |
Individual |
NA185073 |
ILLUMINA, SOLID |
|
Yoruban mother, father, and child trio2 |
NA19238 |
ILLUMINA |
|
|
NA19239 |
ILLUMINA |
||
|
NA19240 |
ILLUMINA, LS454, SOLID |
||
|
East Asian |
Han Chinese individual |
YanHuang (YH) Genome4
|
ILLUMINA |
|
Korean individual |
Seong-Jin Kim (SJK)5 |
ILLUMINA |
Full table of 2,607 superstable elements can be downloaded Table_S1.csv
The chromosomal start position (in hg19 coordinates), end position, length in base pairs, the rank of the length for each superstable elements and identification label are documented in this table.
Superstable elements categorys are listed in Table_S4.xls and Table_S11.xls
Each sheet in Table_S4 spreadsheet lists the respective superstable elements (denoted by chromosome number and length order rank) of each of our 5 classes as detailed in the Methods section. Chromosome name, start position (in hg19 coordinates), end position, identification label, related gene Refseq status and gene name are included in the sheet. In category "unknown-10kb" and "unknown-other", the nearest genes upstream and downstream are offered with gene names and distance.
Each sheet in Table_S11 spreadsheet lists the respective superstable elements (denoted by chromosome number and length order rank) of each of our 27 classes as detailed in the RMethods section and in Supplementary Figure S1.
IV. SNP density Classification
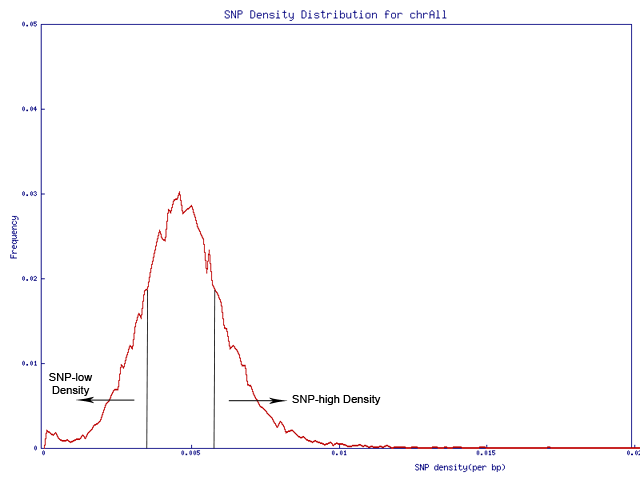
The whole genome is equally split into 3 parts, by calculating SNP density on a 1Mbp window.
Regions with SNP density lower than 0.0041 are classified as SNP-low density regions.
Regions with SNP density higher than 0.0053 are classified as SNP-high density regions.
Other regions are classified as SNP-median density regions.
==================================================
If you have any questions, feel free to contact us.
Email: doodlehzq@sjtu.edu.cn
ccwei@sjtu.edu.cn